An Intro
| ⬅️ Back | Next ➡️ |
A major challenge of any data-related endeavor is the integration of disparate sources of information. German BioImaging (GerBI; https://gerbi-gmb.de/) is currently involved in a project to link all German research outputs at the national level in the National Research Data Infrastructure (NFDI). ΔTissue has solicited GerBI to provide a searchable, linked data graph for the program. Along with our Belgium-based partner, Micelio (https://micelio.be/), we introduce below the linked data framework as well as the proposed steps to bring the same infrastructure to the ΔTissue program.
What is Linked Data?
Linked data builds upon web technologies such as HTTP, RDF and URIs to connect machine-readable representations of research outputs. A very straight-forward introduction can be found at https://ontola.io/what-is-linked-data/ .
The process of formatting one’s data (here we do not differentiate between “data” and “metadata”) into linked data with well-defined identifiers and semantics exposes inconsistencies early in the data acquisition process, leaving one with cleaner, more interoperable graphs of data. Graphs produced from a single research team can then be connected to others through common, matching URIs, providing a single program-spanning, searchable resource. This linked data graph:
- provides a central clearing house for exposing inconsistencies and differences in naming;
- drives the conversation of maximizing the FAIRness of data; and
- serves as a portal through which disparate research areas can communicate
Toy Example
To give a simple, toy example:
| Patient | Observation | Disease State | |
|---|---|---|---|
| Sample#1 | Patient#A | N/A | Early |
| Sample#2 | Patient#A | Elevated count | Late |
| Sample#3 | Patient#B | Prevalent | Post-Mortem |
A typical checklist that one would go through while trying to convert the above spreadsheet into a linked data graph might contain:
- How will the local identifiers in each cell be converted into unique identifiers (URIs)? Is the same scheme used everywhere? Or are the identifiers only unique to the current spreadsheet table?
- What is the exact name of the property represented by the column heading? Again, is there a single URI for all uses of, e.g., “Disease State” or are there slight differences or perhaps restrictions between tables?
- What are the possible values for a given property? Do they come from an existing ontology? Is a new ontology needed?
- Are there any further semantics, limitations that can be captured? Is a given column of a type that is a sub-type or a super-type of another category?
Once this checklist has been applied to all spreadsheets, the table can be written with clickable links (which is of course where the name “Linked Data” comes from).
By using a namespace prefix, we can simplify this text and represent it equivalently as statements in a graph:
@prefix sample: <https://wellcomeleap.org/delta-tissue/samples/>.
@prefix patient: <https://wellcomeleap.org/delta-tissue/patients/>.
@prefix obs: <https://wellcomeleap.org/delta-tissue/observations/>.
sample:10584 sample:acquired-from patient:49602 .
sample:40592 sample:acquired-from patient:49602 .
sample:96049 sample:acquired-from patient:99999 .
sample:96049 obs:observed-with obj:prevalent .
sample:96049 obs:evidence https://wellcomeleap.org/delta-tissue/images/123 .
This process of cleaning the input data, assigning unique global identifiers, and identifying or building ontologies also enables the use of a standardized query language, SPARQL, (https://www.w3.org/TR/2013/REC-sparql11-overview-20130321/):
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX leap: <http://wellcomeleap.org/vocabulary/0.1/>
SELECT ?patientName, ?sampleAge
WHERE {
?patient a leap:Patient .
?patient foaf:name ?patientName .
?sample leap:came_from ?patient .
?sample leap:sample_age ?sampleAge .
}
The sophistication of what is queryable is primarily limited by how well the data is structured. See the example below from Wikidata for biomedical knowledge integration and curation (Gregory Stupp) for a query which “finds drugs for cancers that target genes related to cell proliferation”:
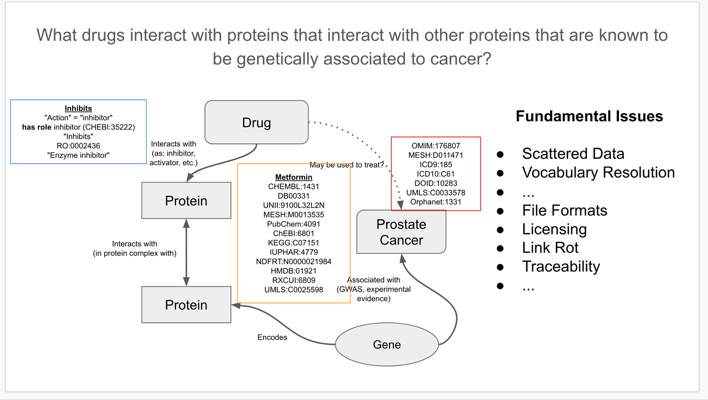
One of the SPARQL queries provided in the slide deck has been copied below as an example. Importantly, though, in the demonstrator the query is then executed against Wikidata and the results are embedded in the page live.
#title: What drugs interact with proteins whose genes are associated with prostate cancer?
#from: "Wikidata for biomedical knowledge integration and curation" by Gregory Stupp (Scripss)
SELECT ?drugLabel ?geneLabel ?diseaseLabel WHERE {
?drug wdt:P129 ?gene_product . # drug interacts with a gene_product
?gene wdt:P688 ?gene_product . # gene_product (usually a protein) is a product of a gene (a region of DNA)
?disease wdt:P2293 ?gene . # genetic association between disease and gene
?disease wdt:P279* wd:Q12078 . # limit to cancers wd:Q12078 (the * operator runs up a transitive relation..)
SERVICE wikibase:label {
bd:serviceParam wikibase:language "en" .
}
}
limit 1000
However, the values returned here are just strings. With linked data, we want to keep the unique
identifiers. The next query returns the objects themselves which you can follow to view the
data entry from where you can explore related concepts.
#title: What drugs interact with proteins whose genes are associated with prostate cancer (Linked)?
#from: "Wikidata for biomedical knowledge integration and curation" by Gregory Stupp (Scripss)
SELECT ?drug ?drugLabel ?gene ?geneLabel ?disease ?diseaseLabel WHERE {
?drug wdt:P129 ?gene_product . # drug interacts with a gene_product
?gene wdt:P688 ?gene_product . # gene_product (usually a protein) is a product of a gene (a region of DNA)
?disease wdt:P2293 ?gene . # genetic association between disease and gene
?disease wdt:P279* wd:Q12078 . # limit to cancers wd:Q12078 (the * operator runs up a transitive relation..)
SERVICE wikibase:label {
bd:serviceParam wikibase:language "en" .
}
}
limit 1000
By clicking on “Wikidata query service”, you can open the query in an editor so that you
can experiment on your own.
Initial project
While the research groups are gathering data, our focus will be on (1) building the linked data platform and (2) discussing the evolving data from the various groups.
Platform
The platform itself will be web-accessible and limited to access by platform participants. A tight integration with any existing and planned platform resources will be investigated but will not be the immediate priority. Instead, the focus will be on accurately capturing cleaned program metadata which will make subsequent transformations or migrations reasonably straight-forward.
There are several platforms that are under investigation. Many of the examples shown in this document are built using Wikidata (https://www.wikidata.org/), probably the most widely known linked data platform. It is, however, more intended for public data but several concrete life sciences graphs have been built on top it, like ChlamBase (https://chlambase.org/). The underlying platform (Wikibase) is a strong candidate, offering editing of individual triples (the basic unit of a graph) and services for processing tabular data.
Alternatively, a pure RDF database like the open-source Virtuoso (https://virtuoso.openlinksw.com/) or the commercial GraphDB (https://www.ontotext.com/products/graphdb/) and Stardog (https://www.stardog.com/) would equally provide the basic needs of storing and searching the graph with tradeoffs on usability. In the end, program participants will minimally be provided the necessary endpoints for graph operations while feedback is solicited on the wider requirements.
Data Specifications
Several groups have begun the process of defining their data schemas. The Subramaniam group, for example, have a minimal clinical data specification that looks to fit the needs of many other groups. From this and similar definitions, the identifiers for the linked data graph can be built.
We would propose kick starting the building process via “Bring-your-own-data” (BYOD) sessions as introduced at SWAT4LS, 2014:
“The basic workflow that we follow in a BYOD is as follows: (i) define a driving user question and demonstrator, (ii) find appropriate ontologies and ontology concepts, (iii) find appropriate source data (data owner data + existing Linked Data), (iv) create Linked Data, (v) create a Linked Data Cache, (vi) perform a SPARQL query that represents the user question. In the first BYOD, driving user questions were chosen on the first day. In the second BYOD defining driving questions and selecting data starts with the preparatory webinars. Participants share information in a Google Document.”
Other examples of BYODs can be found on the “Rare Disease (RD) Connect” webpage, https://rd-connect.eu/?s=%22byod%22 which ran until 2018.
German BioImaging and Micelio will hold preliminary meetings to present the general technologies and process and answer any questions. Dependent on those conversations, a single program-wide BYOD, a per-disease BYOD, or some other combination of sessions will be scheduled. The goal is to identify common terminology and use cases that can be captured in a linked data graph.
Outlook
Throughout the rest of 2022 (Q2-Q4), German BioImaging and Micelio will help the members of ΔTissue to scale up the quantity and quality of linked data in the platform. Further BYOD sessions can be planned as necessary, and the usability of the platform can be improved in collaboration with UX professionals most suited to the chosen platform. We will work to facilitate the entry of data into the platform and/or automate the migration from existing data solutions like Sage Bionetworks.
Most critically, however, concrete queries and use cases for the investigation of the linked data shall be identified and improved over the course of the project. It should be noted that the most value will come from having teams engaging directly with the platform rather than delegating semantic searches to informaticians who have less understanding of the domain.
Terminology
- Graph: or more formally, a labeled, directed, multi-graph, consists of a collection of RDF statements, or triples.
- Linked Data: “LD”, for “large scale integration of, and reasoning on, data on the web”. See https://www.w3.org/standards/semanticweb/data .
- Ontology: For linked data, a vocabulary that defines concepts and relationships which describe and represent an area of concern. In general, “[t]here is no clear division between ‘vocabularies’ and ‘ontologies’” though “the trend is to use the world ‘ontology’ for a more complex, and possibly quite formal collection of terms”. See https://www.w3.org/standards/semanticweb/ontology .
- RDF: “Resource Description Framework”, a standard model of data interchange and a W3C recommendation. See https://www.w3.org/RDF/
- Sparql: specifications for languages and protocols for querying RDF graphs and a W3C recommendation. See https://www.w3.org/TR/2013/REC-sparql11-overview-20130321/
- Triples: The basic unit of an RDF graph consisting of (resource, predicate, object).
- URI: universal resource identifier, a standardized representation of web resources. See https://www.w3.org/Addressing/URL/uri-spec.html
| ⬅️ Back | Next ➡️ |